Unlocking New Frontiers in Drug Discovery: High Throughput Computing in the Thyme Lab
By: Kallen Wank
April 7, 2025
In 2022, computing challenges arose at the Thyme Lab when the supercomputer at the University of Alabama at Birmingham (UAB) couldn’t handle running enough parallel jobs for the Thyme Lab’s large data pipeline. Seeking a solution to this computing bottleneck, Ari Ginsparg, a PhD student working in the lab noted that “a person on my thesis committee recommended the OSG, so we started exploring it. With the OSG, I was able to access more resources — about ten times more — which made a huge difference.”
The Thyme Lab, led by Summer Thyme, PhD, assistant professor of biochemistry & molecular technology, has since moved in 2023 to UMass Chan Medical School, where it continues its work to uncover the molecular basis of complex neurodevelopmental diseases and to develop therapies.
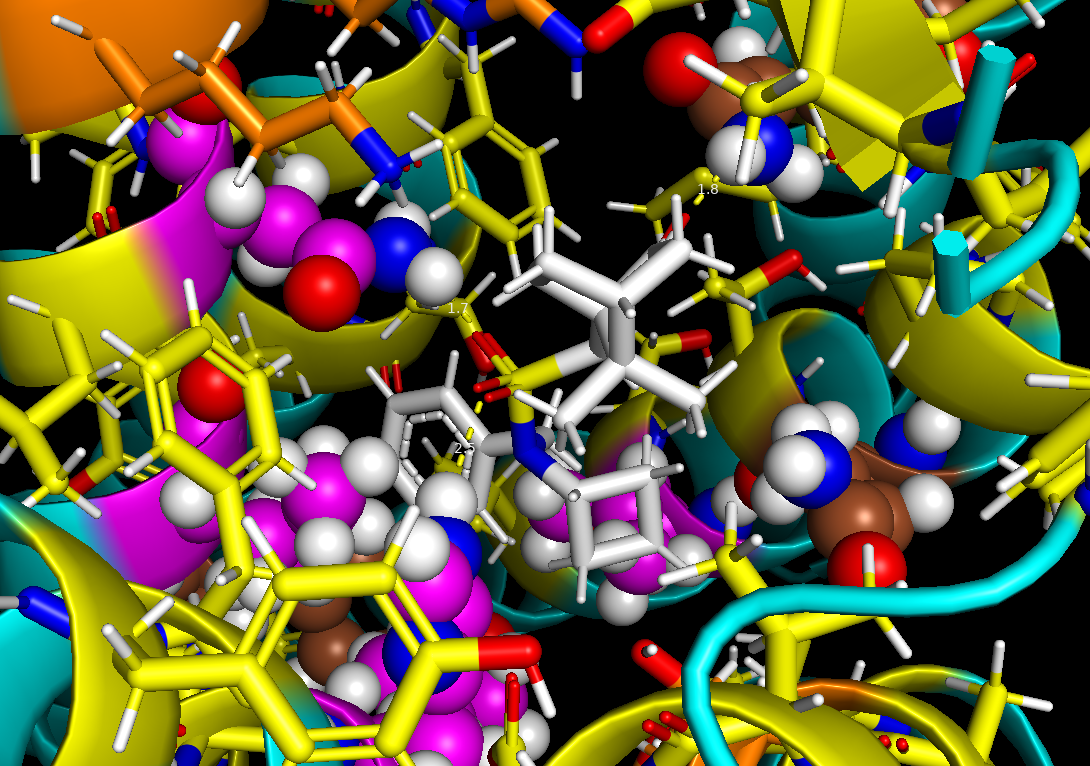
Ginsparg has focused his work on computational drug discovery. His research centers on developing treatments for diseases like narcolepsy and insomnia. With a background in bioinformatics and computational chemistry, he combines computational techniques with experimental biology to design more effective drugs and advance drug discovery for neurodevelopmental disorders.Using Rosetta, a protein modeling software, Ginsparg created an algorithm to predict potential drugs targeting hypocretin receptor 2 (HCRTR2)— a key player in insomnia and narcolepsy.
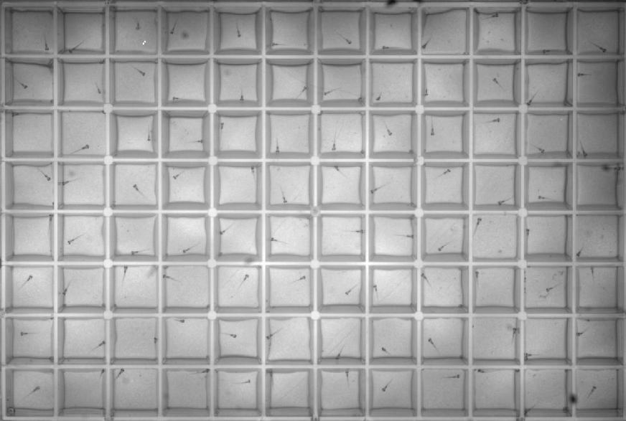
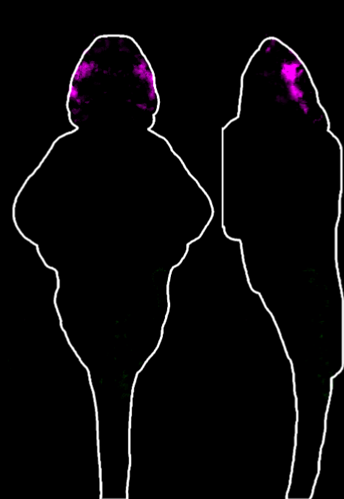
The Thyme Lab uses zebrafish as an ideal model organism to study a wide range of neurodevelopmental disorders. Zebrafish are a popular animal model in labs because they share roughly 70 percent of their DNA with humans, progress through their life cycle more quickly, and are more cost-effective to maintain in the large numbers required for rigorous biochemical studies.
The zebrafish are analyzed with a variety of techniques to view the responses to certain stimuli. For example, Ginsparg explained that “we have specialized devices where zebrafish are placed in 96-well plates and filmed while swimming around. We can add stimuli like flashing lights or sounds to observe how they react.” They also use programs to quantify the sizes of different brain regions and compare them to wild-type zebrafish to see if there are any changes. The data is all uploaded to applications that analyze the images and output results.
To learn how to use the OSPool, a source of computing capacity that is accessible to any researcher affiliated with a US academic institution, Ginsparg relied on the OSG Office Hours. These virtual hours*, led by the Research Computing Facilitators (RCFs), provide guidance and support. When transitioning to OSPool, Ginsparg emphasized “that the office hours and resources from OSG were critical to my success. The amount of resources they provide has been essential for progressing with my thesis project.”
The Thyme lab’s setup to modify their systems to use the OSPool was a gradual process. Ginsparg had already set up a workflow with their local supercomputer, so to adapt the workflow to use the OSPool and its software suite HTCondor, he transitioned to a new software better suited for OSPool.
“We also had to connect the workflow using the OSPool to our data storage system at UAB, which was a bit of a learning process. Once everything was connected, I was able to scale up the workflow,” Ginsparg said. In the Thyme lab, he is the only one currently using OSPool, but commented that it makes a huge difference. In just the past year, the Thyme Lab has run over 4.7 million jobs on the OSPool.
With the jobs running on the OSPool, Ginsparg commented that they have not faced problems or wait times to run jobs. When faced with barriers, Ginsparg returns to OSG Office Hours to get answers to any sort of questions. “The biggest challenge was learning how HTCondor works, but once I got past that, there haven’t been any major issues. The limitations are pretty minimal, and nothing has stopped progress.”
Utilizing the OSPool, Ginsparg, and the Thyme Lab have made discoveries in their search to find new drugs for diseases.“Recently I developed a method to predict drugs that can target a specific pocket in a protein. That’s exciting because it adds new functionality to the Lab’s Rosetta program. We’re on track to develop drugs that could help treat insomnia.” A future direction of the lab includes increasing their usage of OSPool to expand their drug discovery pipeline to identify drugs for other targets.
*OSPool virtual hours occur weekly. Learn more here.